In a recently published article in ACS Nano, we demonstrate a SERS substrate capable of reproducible measurements, such that the relative concentration of a mixture of SERS active molecules can be determined.
Surface enhanced Raman spectroscopy has become a widely used research tool for the identification of molecules, but has yet to achieve widespread use outside of academia. The dominant method for generating the required spectra is to use colloidal metallic nanoparticles, but this leads to an inherently random distribution of SERS-active sites, and thus although the signal can uniquely identify a molecule, the intensity of that signal will vary, rendering quantitative analysis impossible.
We use a lithographically defined fishnet structure, which is uniform across an area of 50 by 50 microns, but also can be fabricated to be identical to other structures on the same host. This allows for direct comparisons between different samples, both qualitatively and quantitatively in terms of the signal intensity.
We then applied this method to detecting the concentration of a potential biomarker for disease, Glycerophosphoinositol (GroPIns), in a physiologically relevant mixture of glycerol and myo-inositol. This could, in the future, allow the analysis for the concentrations of chemical markers of disease straight from cellular fluids themselves, opening the way for new diagnostic methods.
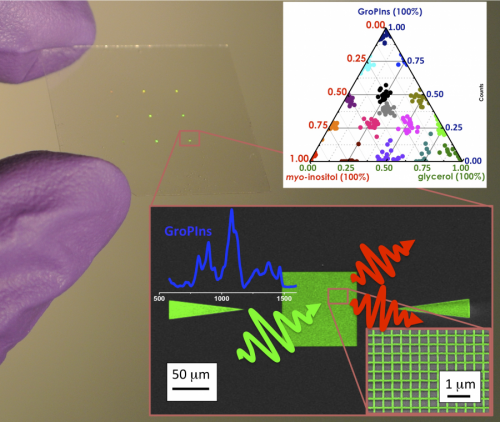
A photograph of the SERS substrate, showing a ternary of the three-component mixtures (top), and an SEM image of the fishnet used